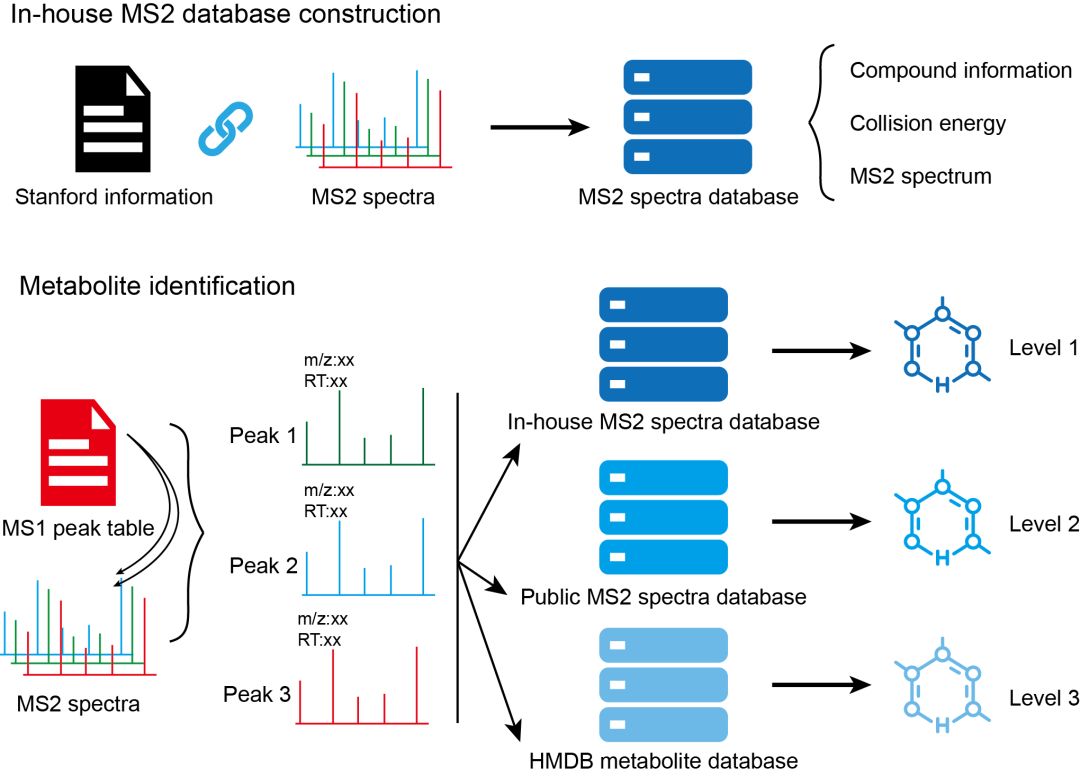
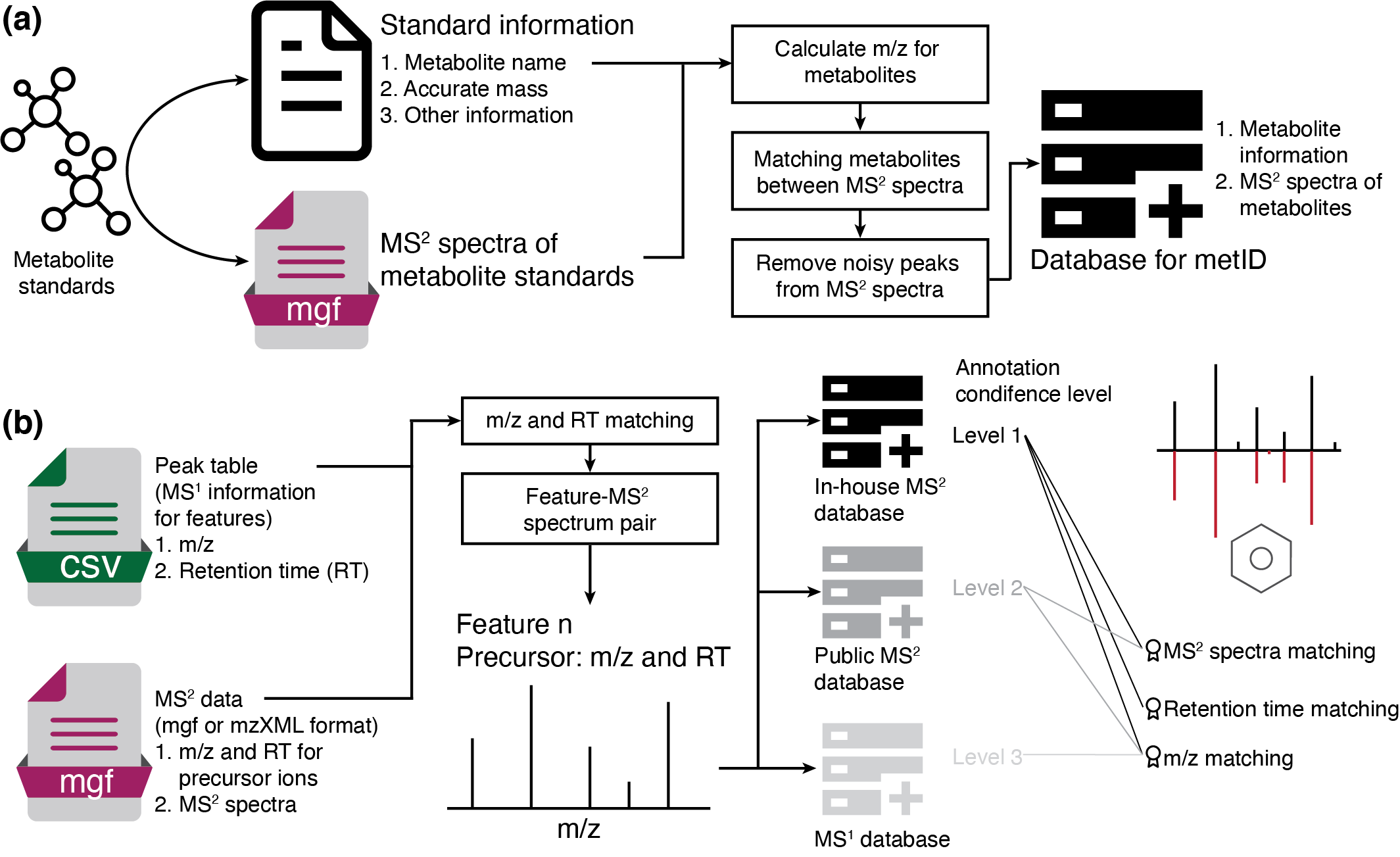
安装 metid包能够用于in-house代谢物库构建,并可利用MS2 spectra进行代谢物鉴定。metid自带数据库,来自于公共数据库的整合。
1 2 3 4 if ( ! require( remotes) ) { install.packages( "remotes" ) } remotes:: install_github( "tidymass/metid" )
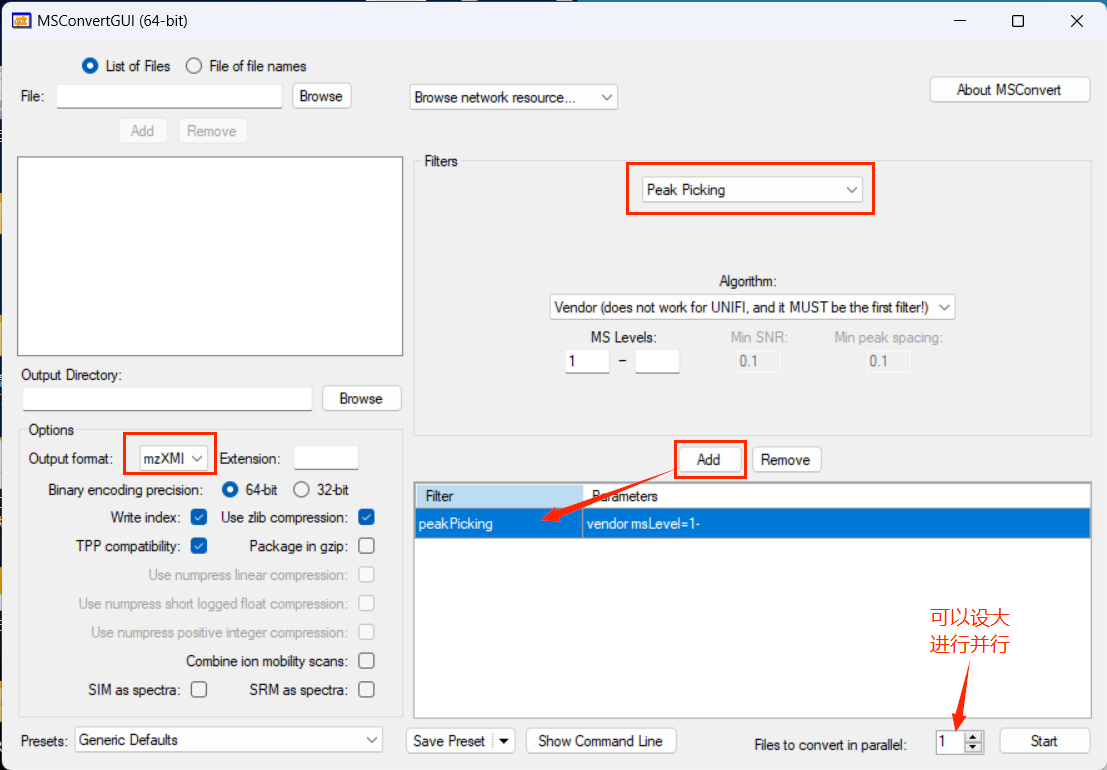
建库 构建内部库 质谱数据准备 将标准品原始质谱数据用ProteoWizard 转换为mzXM格式.
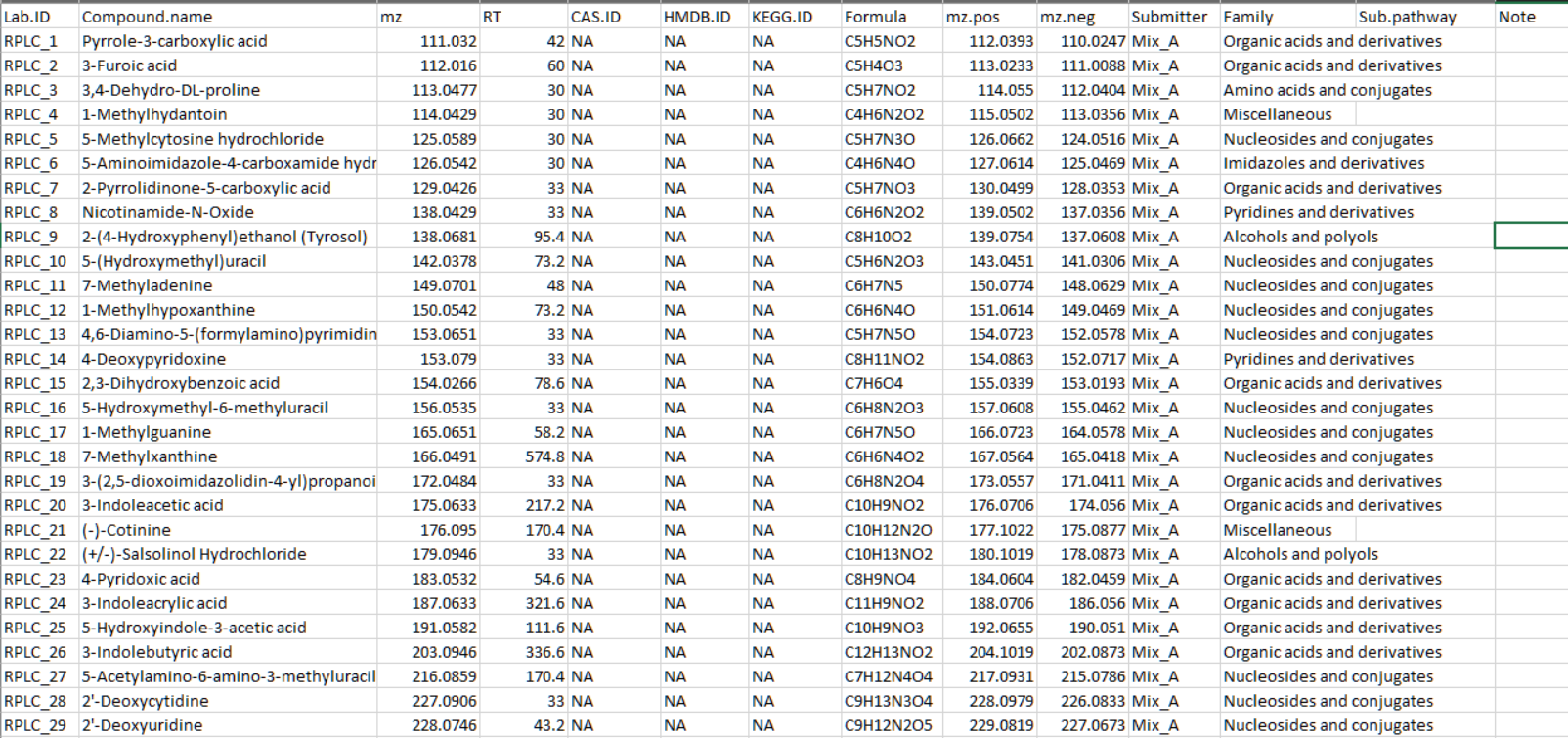
标准品信息表 将标准品信息整理至csv格式表格中,参考如下。
Lab.ID: 不可重复
mz: 化合物准确的mass
RT: 保留时间,以秒为单位
mz.pos: 正离子模式下化合物的mz,如M+H。可设为NA
mz.neg: 负离子模式下化合物的mz,如M-H。可设为NA
Submitter: 个人或组织名称,可设为NA
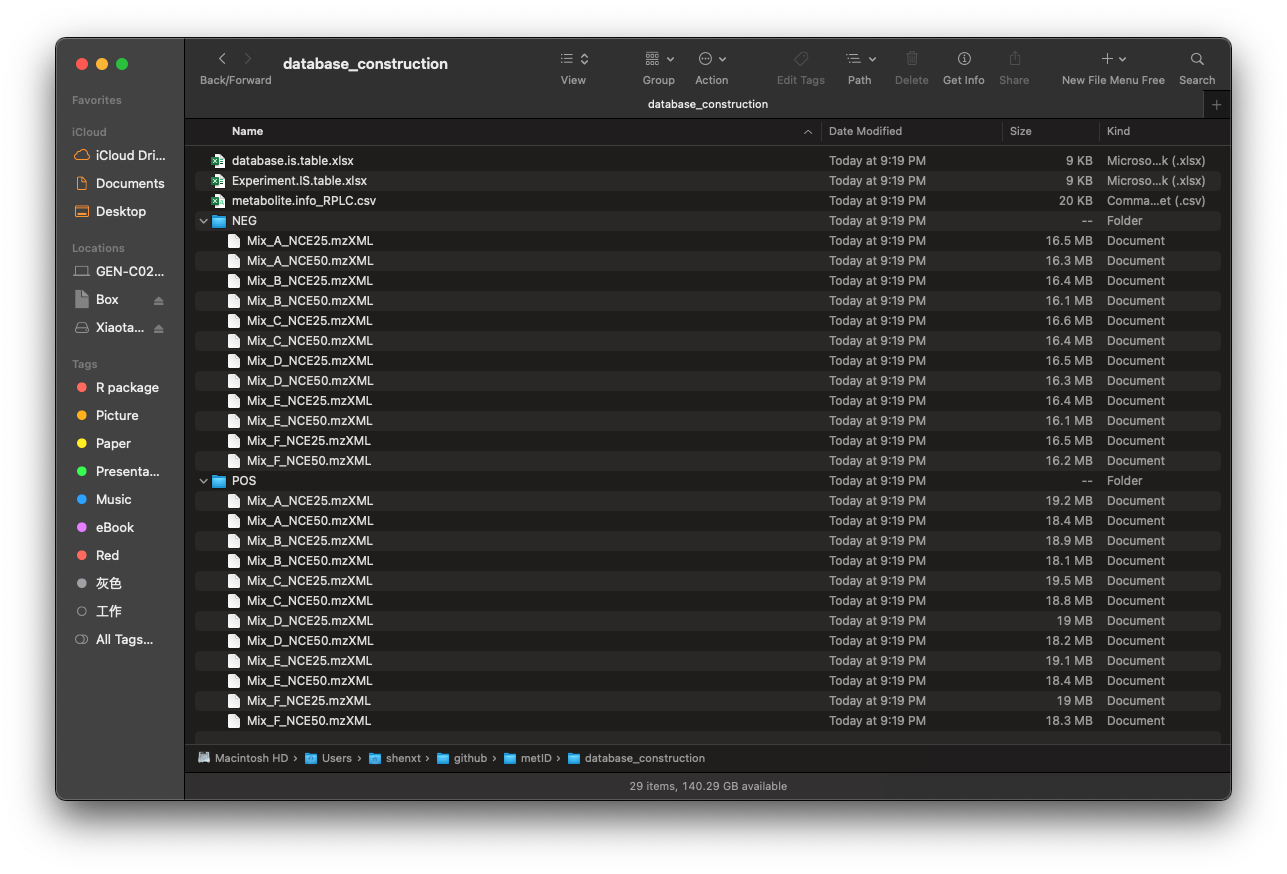
新建目录database_construction,将正离子数据放在database_construction/POS中,将负离子数据放在database_construction/NEG中,标准品信息表metabolite.info_RPLC.csv放在database_construction中。
注意 :每个文件名必须包含碰撞能(collision energy),如test_NCE25.mzXML。
建库 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 library( metid) datapath<- file.path( "./database_construction" ) mydb <- construct_database( path = datapath, version = "0.0.1" , metabolite.info.name = "metabolite.info_RPLC.csv" , source = "my lab" , link = "http://xxx.com" , creater = "someone" , email = "x@126.com" , rt = TRUE , mz.tol = 15 , rt.tol = 30 , threads = 10 ) save( mydb, file= "mydb" )
注意 :保存时前后名字必须一样。
构建公共库 1 remotes:: install_github( "tidymass/massdatabase" )
可将msp格式的数据库转换为metid数据库。目前有bug:Error in `dplyr::select()`: ! Can’t subset columns that don’t exist. ✖ Column `Name` doesn’t exist.
bug解决前可以下载已经构建好的公共库:Database provided for metid 。
下载最新的release MassBank_NIST.msp ,将其放在当前目录下。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 setwd( "C:/Users/liu/Downloads" ) library( metid) massbank_database_2022.12.01 <- construct_mona_database( file = "MassBank_NIST.msp" , path = "." , version = "2022.12.01" , source = "MassBank" , link = "https://github.com/MassBank/MassBank-data/releases" , creater = "Hualin Liu" , email = "LHL371@126.com" , rt = FALSE , threads = 15 ) save( massbank_database_2022.12.01, file = "massbank_database_2022.12.01" )
下载对应数据库,如此处下载LC-MS Spectra (153,242 spectra),截至2023.04.16 。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 library( metid) mona_database_2023.04.16 <- construct_mona_database( file = "MoNA-export-LC-MS_Spectra.msp" , path = "." , version = "2023.04.16" , source = "MoNA" , link = "https://mona.fiehnlab.ucdavis.edu/" , creater = "Hualin Liu" , email = "LHL371@126.com" , rt = FALSE , threads = 10 ) save( mona_database_2023.04.16, file = "mona_database_2023.04.16" ) library( massdatabase) data <- massdatabase:: read_msp_data( "MoNA-export-LC-MS_Spectra.msp" , source = "mona" ) massdatabase:: convert_mona2metid( data = data, path = "." , threads = 10 )
参考
加关注 关注公众号“生信之巅”。
敬告 :使用文中脚本请引用本文网址,请尊重本人的劳动成果,谢谢!Notice : When you use the scripts in this article, please cite the link of this webpage. Thank you!