DOC for V0.0.0
MGCA
Microbial genome component and annotation pipeline
Introduction
The software under designing dedicates to perform the following analysis:
- Genomic Component
- HGT
- Genomic Island
- Prophage
- CRISPR-Cas
- Non-coding RNA
- rRNA
- tRNA
- sRNA
- Repeat Sequences
- Tandem Repeats
- Interspersed Repeats
- HGT
- Genomic Attributes
- Genome Survey
- Protein Properties
- WGS-based Species Identify
- Function Annotation
- General Annotation
- SwissProt
- Pfam
- GO
- KEGG
- Target Gene Mining
- Effectors
- T3SS
- T4SS
- Secretory/Membrane/Intracellular Protein
- Secondary Metabolite Biosynthetic Gene Clusters
- Virulence/Pathogenicity/Resistance Gene
- Antibiotic Resistance Genes (ARGs)
- Pathogen Host Interactions (PHI)
- Comprehensive Antibiotic Resistance Database (CARD)
- Element Cycle
- CAZyme
- Nitrogen
- Sulfur
- Methane
- Membrane Transport Protein (TCDB)
- Effectors
- General Annotation
- Comparative Genomics
- Collinearity
- Positive Selection
- SNP
NOTICE: It will take a long time to complete the development!
Installation
The software was tested successfully on Windows WSL, Linux x64 platform, and macOS. Because this software relies on a large number of other software, so it is recommended to install with Bioconda.
Step1: Install MGCA
-
Method 1: use mamba to install MGCA (
is now avaliable)
# Install mamba first conda install mamba # Usually specify the latest version of PGCGAP mamba create -n mgca mgca=0.0.0
Step2: Setup database (Users should execute this after the first installation of mgca)
conda activate mgca
setupDB --all
conda deactivate
Notice: there is a little bug, users can edit the "setupDB" file located at the mgca installation path to resolve the problem. Just remove the lines after line no. 83.
Required dependencies
- artemis
- bakta
- eggnog-mapper
- emboss
- gtdbtk
- interproscan (include emboss openjdk)
- islandpath
- kakscalculator2
- lastz
- mummer4
- opfi
- Perl & the modules
- phispy
- R & the packages
- repeatmasker
- saspector (include trf progressivemauve prokka)
Usage
-
Print the help messages:
mgca --help -
General usage:
mgca [modules] [options] -
Modules:
-
[--PI] Calculate statistics of protein properties and print pI of all protein sequences
-
[--IS] Predict genomic island from GenBank files
-
[--PROPHAGE] Predict prophage sequences from GenBank files
-
[--CRISPR] Finding CRISPR-Cas systems in genomics or metagenomics datasets
-
Examples
Example 1: Calculate statistics of protein properties and print pI of all protein sequences
mgca --PI --AAsPath <PATH> --aa_suffix <.faa>
Example 2: Predict genomic island from GenBank files
mgca --IS --gbkPath <PATH> --gbk_suffix <.gbk>
Example 3: Predict prophage sequences from GenBank files
mgca --PROPHAGE --gbkPath <PATH> --gbk_suffix <.gbk> --phmms <Path of pVOG.hmm> --phage_genes <1> --min_contig_size <5000> --threads <6>
Example 4: Finding CRISPR-Cas systems in genomics or metagenomics datasets
mgca --CRISPR --scafPath <PATH> --scaf_suffix <.fa> --casDBpath <db path> --threads <6>
OUTPUT
PI
- Results/PI/*.pepstats: Peptide statistics for each protein sequence organized by the genome.
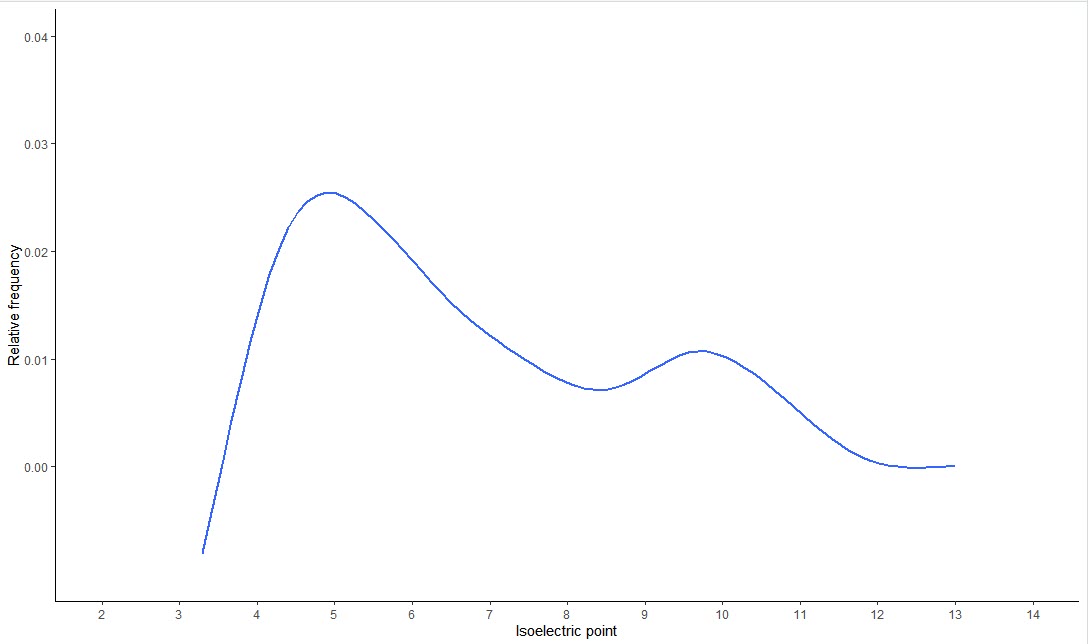
- Results/PI/*.pI: Protein isoelectric point and its frequency.
-
Results/PI/*.pI.tiff: A plot drawing ‘Relative frequency' vs. ‘isoelectric point'.
IS
- Results/IS/All_island.list: A list file containing genomic island information.
| Island IDs | GI Start | GI End | GI Length | Gene Number | Gene IDs |
|---|---|---|---|---|---|
| ID=LHL010_Scaffold2_gi1 | 404015 | 531251 | 127237 | 288 | LHL010_01513 LHL010_01514 LHL010_01515 LHL010_01516 LHL010_01517 LHL010_01518 LHL010_01519 LHL010_01520 LHL010_01521 LHL010_01522 LHL010_01523 LHL010_01524 LHL010_01525 LHL010_01526 LHL010_01527 LHL010_01528 LHL010_01529 LHL010_01530 LHL010_01531 LHL010_01532 LHL010_01533 LHL010_01534 LHL010_01535 LHL010_01536 LHL010_01537 LHL010_01538 LHL010_01539 LHL010_01540 LHL010_01541 LHL010_01542 LHL010_01543 LHL010_01544 LHL010_01545 LHL010_01546 LHL010_01547 LHL010_01548 LHL010_01549 LHL010_01550 LHL010_01551 LHL010_01552 LHL010_01553 LHL010_01554 LHL010_01555 LHL010_01556 LHL010_01557 LHL010_01558 LHL010_01559 LHL010_01560 LHL010_01561 LHL010_01562 LHL010_01563 LHL010_01564 LHL010_01565 LHL010_01566 LHL010_01567 LHL010_01568 LHL010_01569 LHL010_01570 LHL010_01571 LHL010_01572 LHL010_01573 LHL010_01574 LHL010_01575 LHL010_01576 LHL010_01577 LHL010_01578 LHL010_01579 LHL010_01580 LHL010_01581 LHL010_01582 LHL010_01583 LHL010_01584 LHL010_01585 LHL010_01586 LHL010_01587 LHL010_01588 LHL010_01589 LHL010_01590 LHL010_01591 LHL010_01592 LHL010_01593 LHL010_01594 LHL010_01595 LHL010_01596 LHL010_01597 LHL010_01598 LHL010_01599 LHL010_01600 LHL010_01601 LHL010_01602 LHL010_01603 LHL010_01604 LHL010_01605 LHL010_01606 LHL010_01607 LHL010_01608 LHL010_01609 LHL010_01610 LHL010_01611 LHL010_01612 LHL010_01613 LHL010_01614 LHL010_01615 LHL010_01616 LHL010_01617 LHL010_01618 LHL010_01619 LHL010_01620 LHL010_01621 LHL010_01622 LHL010_01623 LHL010_01624 LHL010_01625 LHL010_01626 LHL010_01627 LHL010_01628 LHL010_01629 LHL010_01630 LHL010_01631 LHL010_01632 LHL010_01633 LHL010_01634 LHL010_01635 LHL010_01636 LHL010_01637 LHL010_01638 LHL010_01639 LHL010_01640 LHL010_01641 LHL010_01642 LHL010_01643 LHL010_01644 LHL010_01645 LHL010_01646 LHL010_01647 LHL010_01648 LHL010_01649 LHL010_01650 LHL010_01651 LHL010_01652 LHL010_01653 LHL010_01654 LHL010_01655 LHL010_01656 LHL010_01657 LHL010_01658 LHL010_01659 LHL010_01660 LHL010_01661 LHL010_01662 LHL010_01663 LHL010_01664 LHL010_02694 LHL010_02695 LHL010_02696 LHL010_02697 LHL010_02698 LHL010_02699 LHL010_02700 LHL010_02701 LHL010_02702 LHL010_02703 LHL010_02704 LHL010_02705 LHL010_02706 LHL010_02707 LHL010_02708 LHL010_02709 LHL010_02710 LHL010_02711 LHL010_02712 LHL010_02713 LHL010_02714 LHL010_02715 LHL010_02716 LHL010_02717 LHL010_02718 LHL010_02719 LHL010_02720 LHL010_02721 LHL010_02722 LHL010_02723 LHL010_02724 LHL010_02725 LHL010_02726 LHL010_02727 LHL010_02728 LHL010_02729 LHL010_02730 LHL010_02731 LHL010_02732 LHL010_02733 LHL010_02734 LHL010_02735 LHL010_02736 LHL010_02737 LHL010_02738 LHL010_02739 LHL010_02740 LHL010_02741 LHL010_02742 LHL010_02743 LHL010_02744 LHL010_02745 LHL010_02746 LHL010_02747 LHL010_02748 LHL010_02749 LHL010_02750 LHL010_02751 LHL010_02752 LHL010_02753 LHL010_02754 LHL010_02755 LHL010_02756 LHL010_02757 LHL010_02758 LHL010_02759 LHL010_02760 LHL010_02761 LHL010_02762 LHL010_02763 LHL010_02764 LHL010_02765 LHL010_02766 LHL010_02767 LHL010_02768 LHL010_02769 LHL010_02770 LHL010_02771 LHL010_02772 LHL010_02773 LHL010_02774 LHL010_02775 LHL010_02776 LHL010_02777 LHL010_02778 LHL010_02779 LHL010_02780 LHL010_02781 LHL010_02782 LHL010_02783 LHL010_02784 LHL010_02785 LHL010_02786 LHL010_02787 LHL010_02788 LHL010_02789 LHL010_02790 LHL010_02791 LHL010_02792 LHL010_02793 LHL010_02794 LHL010_02795 LHL010_02796 LHL010_02797 LHL010_02798 LHL010_02799 LHL010_02800 LHL010_02801 LHL010_02802 LHL010_02803 LHL010_02804 LHL010_02805 LHL010_02806 LHL010_02807 LHL010_02808 LHL010_02809 LHL010_02810 LHL010_02811 LHL010_02812 LHL010_02813 LHL010_02814 LHL010_02815 LHL010_02816 LHL010_02817 LHL010_02818 LHL010_02819 LHL010_02820 LHL010_02821 LHL010_02822 LHL010_02823 LHL010_02824 LHL010_02825 LHL010_02826 LHL010_02827 LHL010_02828 LHL010_02829 |
| ID=LHL010_Scaffold2_gi2 | 965322 | 993880 | 28559 | 65 | LHL010_02163 LHL010_02164 LHL010_02165 LHL010_02166 LHL010_02167 LHL010_02168 LHL010_02169 LHL010_02170 LHL010_02171 LHL010_02172 LHL010_02173 LHL010_02174 LHL010_02175 LHL010_02176 LHL010_02177 LHL010_02178 LHL010_02179 LHL010_02180 LHL010_02181 LHL010_02182 LHL010_02183 LHL010_02184 LHL010_02185 LHL010_02186 LHL010_02187 LHL010_02188 LHL010_02189 LHL010_02190 LHL010_02191 LHL010_02192 LHL010_02193 LHL010_02194 LHL010_02195 LHL010_02196 LHL010_03213 LHL010_03214 LHL010_03215 LHL010_03216 LHL010_03217 LHL010_03218 LHL010_03219 LHL010_03220 LHL010_03221 LHL010_03222 LHL010_03223 LHL010_03224 LHL010_03225 LHL010_03226 LHL010_03227 LHL010_03228 LHL010_03229 LHL010_03230 LHL010_03231 LHL010_03232 LHL010_03233 LHL010_03234 LHL010_03235 LHL010_03236 LHL010_03237 LHL010_03238 LHL010_03239 LHL010_03240 LHL010_03241 LHL010_03242 LHL010_03243 |
| ID=LHL010_Scaffold3_gi1 | 371817 | 389128 | 17312 | 22 | LHL010_02659 LHL010_02660 LHL010_02661 LHL010_02662 LHL010_02663 LHL010_02664 LHL010_02665 LHL010_02666 LHL010_02667 LHL010_02668 LHL010_02669 LHL010_02670 LHL010_02671 LHL010_02672 LHL010_02673 LHL010_02674 LHL010_02675 LHL010_02676 LHL010_02677 LHL010_02678 LHL010_02679 LHL010_02680 |
- Results/IS/All_island.txt: A file contains information and sequence of genes in the genomic island.
| Sequence IDs | Predictor | Category | GI Start | GI End | . | Strand | . | Island IDs | Gene IDs | Gene Start | Gene End | Strand | Products | Protein Sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LHL010_Scaffold2 | islandpath | genomic_island | 404015 | 531251 | . | - | . | ID=LHL010_Scaffold2_gi1 | LHL010_01513 | 404015 | 404314 | -1 | hypothetical protein | MNPIIEYLTGMNVLTDQIIAMDLLISAKNGVRNYAMAATEAGTPEVKEVLIRHLEEALDMHEQLSSYMMEKGWYHPWNTDEQVKLHLKNIDTAIQLPTL |
| LHL010_Scaffold2 | islandpath | genomic_island | 404015 | 531251 | . | - | . | ID=LHL010_Scaffold2_gi1 | LHL010_01514 | 404311 | 404526 | -1 | hypothetical protein | MEYALHELLEVQEMTAFKTLCLTKSKTMKALVSDPKLKEIMQQDVDITTRQLQEFASILSNAKQVEEMSEK |
| LHL010_Scaffold2 | islandpath | genomic_island | 404015 | 531251 | . | - | . | ID=LHL010_Scaffold2_gi1 | LHL010_01515 | 404545 | 405681 | -1 | putative zinc-binding alcohol dehydrogenase | MKAVTYQGIKNVVVKDVPDPKIEKSDDMIIKVTSTAICGSDLHLIHGFIPNLQEDYVIGHEPMGIVEEVGPGVTKVKKGDRVIIPFTIACGECFFCKNQLESQCDQSNENGEMGAYFGYSGQTGGYPGGQAEYLRVPFANFTHFKIPESCEEPDEKLSVIADAMTTGFWSVDNAGVKEGDTVIVLGCGPVGLFAQKFCWLKGAKRVIAVDYVNYRLQHAKRTNKVEIVNFEDHENTGNYLKEITKGGADVVIDAVGMDGKMSDLEFLASGLKLHGGTMSALVIASQAVRKGGTIQITGVYGGRYNGFPLGDIMQRNINIRSGQAPVIHYMPYMFELVSTGKIDPGDVVSHVLPLSEAKHGYDIFDAKMDDCIKVVLKP |
PROPHAGE
- Results/PROPHAGE/*_prophage: Result for each genome.
- Results/PROPHAGE/All.prophages.txt: The summary results (for all genomes) include information of prophage on the host genome.
| Strain | Phage ID | Contig ID | Start location of the prophage | Stop location of the prophage | Start of attL | End of attL | Start of attR | End of attR | sequence of attL | Sequence of attR | Why this att site was chosen for this prophage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F06_bin.1 | pp1 | NODE_1250 | 268 | 6260 | 552 | 565 | 3084 | 3097 | CTTCTTCGGCTGG | CCAGCCGAAGAAG | Longest Repeat flanking phage and within 2000 bp |
| LHL010 | pp1 | Scaffold2 | 478039 | 506323 | 476381 | 476394 | 507152 | 507165 | AAAAACAACATCC | GGATGTTGTTTTT | Longest Repeat flanking phage and within 2000 bp |
| LHL010 | pp2 | Scaffold3 | 364543 | 381309 | 364452 | 364464 | 378016 | 378028 | ACGGAAAGCCTG | CAGGCTTTCCGT | Longest Repeat flanking phage and within 2000 bp |
- Results/PROPHAGE/All.prophages.seq: The summary results (for all genomes) include information of prophage genes and sequences.
| Strain | Phage ID | Contig ID | Gene Start | Gene End | Strand | Annotation | Protein sequences |
|---|---|---|---|---|---|---|---|
| F06_bin.1 | pp1 | NODE_1250 | 268 | 1167 | 1 | Tyrosine recombinase XerC | MSVEQLPLFSLIRESPLSAAISGFHEHMIRKGFTENTIKAFLNDLRILTRYLGDDRALSQIGTSELKDFMTYLRHYRGVPCNPKSYARRMTTLKVFFGWLAEEGIIPSDPAAPLIHQRASTPLPQILYAGQVEKLLQATEGLMHAEKPDARPHLLVTFLLQTGIKKGECMALQLNDIDTSEPKAPVLYIRYSNPKMKHKERKLRLSVDFVPILRQYLAEYQPQERLFECTARNLEYVLDNAARLAGLETISFEILRWTCAVRDYKAKMPSDKLRQKLGLSRITWQEASEKLKKLASPPL |
| F06_bin.1 | pp1 | NODE_1250 | 1297 | 1923 | -1 | hypothetical protein | MGIRDLFKPREDKFMRLLVEQAAKTVEGMKLLEEFMEVTDREVAKRLVQVEKEADEVRRILIDELNRSFITPIDREDIFALSLTIDDILDYGYSTVDEMVILEVESNSYLRRMVSLVREAANEIYMGVLRLQDHPNVANDHAVRAKALENRVETVYREALAELFHGPTDIDHIMEMLKLREIYRHLSNAADRGDQAANVIGDIVVKMT |
| F06_bin.1 | pp1 | NODE_1250 | 1947 | 2936 | -1 | hypothetical protein | MNLPSVGIAIIGVALFSDFINGFHGSSNVVATMISSRAMSARNALALSAVAHFCGPFLFGVAVATTIGHEVVDPKAITIVVIFAALLAAIIWNIITWAFGIPSSSFHALVGGLIGAVSVDFGFAMIQMRGLLKVIIALFISPILGLVAGYLLMRLVLFLARGASPRINWFFKRAQIITSLALALGHGTNDAQKTMGIIAMALVTIGYSSEFSVPWWVVALCAGAMSLGTAFGGWRIIRTLGGKFYKIRPVHGFTAQVASAFVILGAALLGGPVSTTQVVSSAILGVGSAKRVSRVRWNVVGNIVAAWVLTIPASALLAALLHLLMRSLL |
| F06_bin.1 | pp1 | NODE_1250 | 2933 | 3109 | -1 | hypothetical protein | MWEEASSAGLRLLTGDALCVKVGTTWGQSQVIEAYANLREDERFTPGVENRSTSEGES |
| F06_bin.1 | pp1 | NODE_1250 | 3122 | 4144 | -1 | hypothetical protein | MSTEVIVAIGGGSNIDAAKAAGVLAILGGPISNYFGTGRVTAFLARGFGLEGLSPKARKSLTPLVAVQTASSSGAHLTKYSNVTDLKTGQKKLIVDEAIIPPRAVFQYDVTTSMPPGFTADGALDGIAHLVEVLLGAVGKGFYQRVEEVARVGIRLIVNHLEQAVAEPTNLEAREAVGLGTDLGGYAIMIGGTSGGHLTSFSLVDILSHGRACAIMNPYYIVFFAPAIEEPLKLIGKIYQEAGLTEASIEALSGRGLGTAVAEAMIALSERIGLPTTLGEVQGFTDGHIERALRAAKDPQLKMKLQNMPIPLTAEMVDEYMRPLLEAARDGDLTLIKNVV |
| F06_bin.1 | pp1 | NODE_1250 | 4141 | 4428 | -1 | hypothetical protein | MEFNVFERARDLLREFKGDSYAFGAGALARTGELASQLGQRAAIIVPSHPGSEGISSQVERSLESAGVVVVGEIQGARPNAPREDVSGWQASFKS |
| F06_bin.1 | pp1 | NODE_1250 | 4436 | 4807 | -1 | hypothetical protein | MPRIKITAGDVVAFAELNETATAEAIWNALPIEARVNTWGDEIYFSIPVYLEEENAQAIVEKGDLGYWPSGNAFCIFFGRTPASRGDEIRPASPVNVFGHLEGDPTIFKSVRSGEKVMLERVE |
| F06_bin.1 | pp1 | NODE_1250 | 4800 | 5174 | -1 | 5-keto-L-gluconate epimerase | MRLALEPINRYETTLINNVAQGLELVERAAADNMGLLLDTFHMNIEEPSIEESIRAAGERLFHFHVADSNRWYPGAGHLDFARILAVLREIGYEGYLSAEILSLPDADTCARRTIEFLKGVFDA |
| F06_bin.1 | pp1 | NODE_1250 | 5205 | 5636 | -1 | 5-keto-L-gluconate epimerase | MKLSIVLSTQPAQFEAIAFKGGFERNVARIAELGYDGVELAIRDPNLVDANGMLRVVSAHGLEVPAIGTGQAWGEEGLSFTDPDRAIRAAAIERTKSHIPFATRTGAACGEHGRTVIIIGLLRGIVKPGVDHAQAMDWLVDAL |
| F06_bin.1 | pp1 | NODE_1250 | 5709 | 6260 | -1 | hypothetical protein | MVLETGKGQGVDQALAEATGGCLQLKVSGIYQEILWRVLAESPDPAERALFEKAWAHTYRIAETLSRVYVALIAGRDQAEAQALLADESRVGKVAGREAIALVKQTLGYGLHQAALAHDLLGVADSSHRHPADDFFRHLSYLAFRPLRAELYRTITPDGWQHYADAVTRYTRMRIAGLGMTED |
CRISPR
- Results/CRISPR/*_intially: Results obtained by permissive BLAST parameters (In most cases, it can be ignored).
- Results/CRISPR/*_filtered: The results obtained after
*_intiallyquality control (The final result). - Results/CRISPR/*_filtered/*.csv: The file contains information of
CRISPR array, as shown below:
| Contig | Loc_coordinates | Name | Coordinates | ORFID | Strand | Accession | E_val | Description | Sequence | Bitscore | Rawscore | Aln_len | Pident | Nident | Mismatch | Positive | Gapopen | Gaps | Ppos | Qcovhsp | Contig_filename |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 338499..363015 | cas1 | 352041..348499 | lcl|352041|348499|2|-1 | -1 | UniRef50_A0A0G3CU52 | 0.00E+00 | UniRef50_A0A0G3CU52 cas1 CRISPR-associated protein Cas3 n=238 RepID=A0A0G3CU52_9GAMM | MIVTFVSQCEKYALKRTRRVLDAFANRIGDNTWQTLITAEGLETVKKMLRKSASRSTAVSCHWIRSRSRTQLLWVVGNRKKFNSEGYVPVNRTEKSFLGREHENDWQYLSLIEAFAKLAALLHDWGKASRLFQDKLNPEIKTPFKGDPVRHEWVSSILFSALVKSQPNSENDENWLGVLSTQQWDESQLQQWAKQNQQAGLPLSGLPDAASLLTWLILSHHRLPFLDVKQQQGDAKNGDRLLLSQIGHKEANTLADMMTFIEQYWGYENKYHEQEFNQRLGMCFEFPNGLLSTSPIWTQAIKQAAIHLKQQLPLFQQAMADGSWKIIAHYARLCLMLGDHNYSSKENDPQWKSDIPLYANTHYVKENPLNKGMQFKQKLDEHLVKVAEIARDVAEQLPFFEGEPPTAGEVKKLTHQKNSTGPFSWQEDVVSTIHKYREQQDDIKGYFIVNMASTGCGKTRANAKVMQALSEDKQSLRFILALGLRTLTLQTGDEYKDSIGLEEDELAVLVGSKAVTQLHHHAADADNSAKDDDNAENKTKPQDIGSESQETLFDDDEEVLLWQEDKWQGALPEEALSTVLTKAKDRALLYAPVLACTIDHIMGATETTRGGRYILPSLRLMSSDLVIDEVDDFSDDDLIAIGRLIHFAGLLGRKVMISSATIPPDTALAYFHAYQSGWYIHAKSRQQPLQIGCIWMDEGKYNSAKDQCQATTQIATITANDVNSVYLYQQYHDQFVDKRAEVLQSLPARRKALIVPMPIEQGKNSQKRFFEHIQAAILTQHEHHHFVDQQTGIQLSFGVVRVANISPCVELTRFLLECEWPENIEIRTMAYHSRQVLLLRHEQEQHLDKVLKRKEKMGEQPIALADPIIRQHLTETQGKAKNLIFILVATPVEEVGRDHDFDWAVVEPSSYRSIIQVSGRVRRHREGEVEQPNVSLLQYNWKGFQGKEKYVFVRPGYENSAIKLKTHNLSELVDEQQLRKSINSLPRIKKPTNVEGTKSCNFASLEHASISYTLKTNCLFPTEQVGKTVKTHQRRRSRANSSAMVRVNTASHLWGYTHGCWWMTAIPVHLAPFRKNSLSIRLSLVISNRGKPEFWEYDRVSGWVTKDSVSGVQYVDFPKSLKERLWLVRDYQQSIYQHSQDEGQLTSISRLYGEVSFLKRDNDNHNYQYDDQMGIYEVER | 1208 | 3126 | 1181 | 54.022 | 638 | 476 | 810 | 20 | 67 | 68.59 | 99 | Contig.fasta |
| 3 | 338499..363015 | cas1 | 353015..352038 | lcl|353015|352038|3|-1 | -1 | UniRef50_A0A1A8T6S6 | 0.00E+00 | UniRef50_A0A1A8T6S6 cas1 CRISPR-associated endonuclease Cas1 n=712 RepID=A0A1A8T6S6_9GAMM | MDDFSPSDLKTILHSKRANMYYLEHCRVMQKDGRVLYLTEAKNENLYFNIPIANTTVLMLGTGTSITQAAMRMLSQAGVLVGFCGGGGTPLYMASEVEWLTPQSEYRPTEYLQGWMKFWFDDTERLRAAKSLQRARIKYLQKIWQNSRELQNEGFIYNDSLIQSALDTFQIRTDTATNSQELLLTEAQLTKNLYKYAANNTGQKHFSRQHKSIDKANAFLNHGNYLAYGLAASCLWVLGIPHGFAVMHGKTRRGALVFDIADLIKDAIVLPWSFICAKEDASEQEFRQQILQSFVDNHALDYLFDTVKAIALQTGSEQQTQEPTQ | 516 | 1328 | 326 | 73.62 | 240 | 81 | 273 | 1 | 5 | 83.74 | 99 | Contig.fasta |
| 3 | 338499..363015 | cas6 | 345030..344389 | lcl|345030|344389|2|-1 | -1 | UniRef50_B2PYF6 | 3.16E-116 | UniRef50_B2PYF6 cas6 CRISPR-derived RNA endonuclease Csy4 n=177 Tax=Bacteria TaxID=2 RepID=B2PYF6_PROST | MNYYQEITLLPDPTIPLDFLWQKVYQQTHIALVDNKSAAGDSAVAIAFPEYGSVGFRLGKKMRLLAKTEQALVQLNISRWLERLSDYVHIKSIQLVPEYATAVSYVRQHVKGEKRIQLDMQKKARLYATKSGLSVEACLAQLKEKQPKAQSRLPFLWVESQQTKSRNEASGHRPFPLFIKCLSAEKPQTGLFNCYGLSQAVTGDIKLATVPHF | 328 | 842 | 213 | 71.362 | 152 | 59 | 182 | 1 | 2 | 85.45 | 100 | Contig.fasta |
| 3 | 338499..363015 | cas7 | 346064..345039 | lcl|346064|345039|3|-1 | -1 | UniRef50_A0A2C5THW8 | 1.95E-122 | UniRef50_A0A2C5THW8 cas7 Type I-F CRISPR-associated protein Cas7f/Csy3 n=32 Tax=Enterobacterales TaxID=91347 RepID=A0A2C5THW8_MORMO | MAKNNDVASVLAFEKKLVPSDGYFYGTSWDNKSQFTPLALQEKSVRGTISNRLKGAVKNDPLKLNAEVEKANLQTVDACALGTEQDTLKHQFTLKVLGGVESPSACNNALFKESYTKAAKEYIEKEGFRELGRRYAHNIANARFLWRNRVGAEKIEVEVNILNSGQEQQWTFDATQYSIRSFDKQDKQVTELGNKIASALANSEGALLLEITTYAQLGKAQEVYPSEELVMDKGKGNKSKILYHVNGHAAMHSQKVGNALRSIDTWYPAYDDEVNSAGAIAIEPYGAVTNLGTAYRTPGAKQDFYTHFDKWARGEKLARVEDEHYVMAVLVRGGVFGESDK | 355 | 910 | 332 | 50.904 | 169 | 161 | 229 | 2 | 2 | 68.98 | 97 | Contig.fasta |
| 3 | 338499..363015 | cas5 | 346999..346067 | lcl|346999|346067|1|-1 | -1 | UniRef50_B2PYF4 | 2.08E-162 | UniRef50_B2PYF4 cas5 CRISPR-associated protein, Csy2 family n=61 Tax=Enterobacterales TaxID=91347 RepID=B2PYF4_PROST | MSEPIYLIKLPHLKVLNANALSSPLTIGFPAMTAWLGFMHALERKLNNDYELDVQFDALAVISHECNLQTYRGPGDFVNSIIGTGNPLDKDGNRSAFIEEARCHLDVSLLIKYEDNEDSPINTAVLNKIHELIPTMKLAGGDILSVEMPQRYILPDGYNNAEKIKLFNSLMPGYALIERRDLMMEAMQEGQDAMDALLDYVTVNNQCVEIENSDESKKKPFEWKMQRKTSGWIVPIATGFQGISPLGKAKNQRDPECSHRFAESMITLGQFLMVNKAECPDEILWCYAQDFENNLYLCEQVQPKHFTSGE | 454 | 1167 | 315 | 68.889 | 217 | 93 | 264 | 2 | 5 | 83.81 | 100 | Contig.fasta |
| 3 | 338499..363015 | cas8 | 348267..346996 | lcl|348267|346996|2|-1 | -1 | UniRef50_A1SUQ0 | 3.98E-162 | UniRef50_A1SUQ0 cas8 CRISPR-associated protein, Csy1 family n=55 Tax=Proteobacteria TaxID=1224 RepID=A1SUQ0_PSYIN | MLDPAIDAFFAERKEGWLKKNLKASMNEHEVSQLQQECEKTFSLNEWLPSAAKRAGQISISTHPCTFSHPSARKNKNGSVSATIAHSQYANDGFLRSGNVTVEADALGNAAALDVYKFLTLQMQDHKTLLSHIEAETPLAKSLLTINTGSYQELREGFLAMAATDETAVTSSKIKQVYFPVWTDHEDYHLLSVLTPSGIVFEMRRRIDNIRFSEQTKALRDLKRKGEYSEMGFKEIYGITTIGFGGTKPQNISVLNNQNAGKAHLLPSLPPVINIRETRLPSKNFFADCINPWHIKEAFEAFHGLITLPKDKRGSRFSEYRDNRIQEYVDHIIIMMWKIRRAYEQEDVVLPTKLAKYQQTWLFPNMQQQRDDLDDWLLVLIAEITRQFIAGYNKVNGKKALSFGNDEFNAIAEIVAKNKELLR | 462 | 1188 | 428 | 54.673 | 234 | 179 | 302 | 5 | 15 | 70.56 | 100 | Contig.fasta |
| 3 | 338499..363015 | cas3 | 352041..348499 | lcl|352041|348499|2|-1 | -1 | UniRef50_A0A0G3CU52 | 0.00E+00 | UniRef50_A0A0G3CU52 cas3 CRISPR-associated protein Cas3 n=238 RepID=A0A0G3CU52_9GAMM | MIVTFVSQCEKYALKRTRRVLDAFANRIGDNTWQTLITAEGLETVKKMLRKSASRSTAVSCHWIRSRSRTQLLWVVGNRKKFNSEGYVPVNRTEKSFLGREHENDWQYLSLIEAFAKLAALLHDWGKASRLFQDKLNPEIKTPFKGDPVRHEWVSSILFSALVKSQPNSENDENWLGVLSTQQWDESQLQQWAKQNQQAGLPLSGLPDAASLLTWLILSHHRLPFLDVKQQQGDAKNGDRLLLSQIGHKEANTLADMMTFIEQYWGYENKYHEQEFNQRLGMCFEFPNGLLSTSPIWTQAIKQAAIHLKQQLPLFQQAMADGSWKIIAHYARLCLMLGDHNYSSKENDPQWKSDIPLYANTHYVKENPLNKGMQFKQKLDEHLVKVAEIARDVAEQLPFFEGEPPTAGEVKKLTHQKNSTGPFSWQEDVVSTIHKYREQQDDIKGYFIVNMASTGCGKTRANAKVMQALSEDKQSLRFILALGLRTLTLQTGDEYKDSIGLEEDELAVLVGSKAVTQLHHHAADADNSAKDDDNAENKTKPQDIGSESQETLFDDDEEVLLWQEDKWQGALPEEALSTVLTKAKDRALLYAPVLACTIDHIMGATETTRGGRYILPSLRLMSSDLVIDEVDDFSDDDLIAIGRLIHFAGLLGRKVMISSATIPPDTALAYFHAYQSGWYIHAKSRQQPLQIGCIWMDEGKYNSAKDQCQATTQIATITANDVNSVYLYQQYHDQFVDKRAEVLQSLPARRKALIVPMPIEQGKNSQKRFFEHIQAAILTQHEHHHFVDQQTGIQLSFGVVRVANISPCVELTRFLLECEWPENIEIRTMAYHSRQVLLLRHEQEQHLDKVLKRKEKMGEQPIALADPIIRQHLTETQGKAKNLIFILVATPVEEVGRDHDFDWAVVEPSSYRSIIQVSGRVRRHREGEVEQPNVSLLQYNWKGFQGKEKYVFVRPGYENSAIKLKTHNLSELVDEQQLRKSINSLPRIKKPTNVEGTKSCNFASLEHASISYTLKTNCLFPTEQVGKTVKTHQRRRSRANSSAMVRVNTASHLWGYTHGCWWMTAIPVHLAPFRKNSLSIRLSLVISNRGKPEFWEYDRVSGWVTKDSVSGVQYVDFPKSLKERLWLVRDYQQSIYQHSQDEGQLTSISRLYGEVSFLKRDNDNHNYQYDDQMGIYEVER | 1208 | 3126 | 1181 | 54.022 | 638 | 476 | 810 | 20 | 67 | 68.59 | 99 | Contig.fasta |
| 3 | 338499..363015 | cas7 | 353015..352038 | lcl|353015|352038|3|-1 | -1 | UniRef50_A0A2G6NPC1 | 4.70E-68 | UniRef50_A0A2G6NPC1 cas7 Subtype I-F CRISPR-associated endonuclease Cas1 (Fragment) n=3 Tax=Proteobacteria TaxID=1224 RepID=A0A2G6NPC1_9DELT | MDDFSPSDLKTILHSKRANMYYLEHCRVMQKDGRVLYLTEAKNENLYFNIPIANTTVLMLGTGTSITQAAMRMLSQAGVLVGFCGGGGTPLYMASEVEWLTPQSEYRPTEYLQGWMKFWFDDTERLRAAKSLQRARIKYLQKIWQNSRELQNEGFIYNDSLIQSALDTFQIRTDTATNSQELLLTEAQLTKNLYKYAANNTGQKHFSRQHKSIDKANAFLNHGNYLAYGLAASCLWVLGIPHGFAVMHGKTRRGALVFDIADLIKDAIVLPWSFICAKEDASEQEFRQQILQSFVDNHALDYLFDTVKAIALQTGSEQQTQEPTQ | 207 | 528 | 122 | 75.41 | 92 | 28 | 110 | 1 | 2 | 90.16 | 38 | Contig.fasta |
| 3 | 338499..363015 | CRISPR array | 343210..344259 | Copies: 18, Repeat: 28, Spacer: 32 | TTTCTAAGCTGCCTGTGCGGCAGTGAAC | Contig.fasta | |||||||||||||||
| 3 | 338499..363015 | CRISPR array | 353308..354476 | Copies: 20, Repeat: 28, Spacer: 32 | TTTCTAAGCTGCCTATACGGCAGTGAAC | Contig.fasta |
Descriptions of each output field are provided below:
| Field name | Description |
|---|---|
| Contig | ID/accession for the parent contig/genome sequence. |
| Loc_coordinates | Start and end position of the candidate locus (relative to the parent sequence). |
| Name | Feature name/label. This is will be identical to "Description" (index 8) if parse_descriptions is True. |
| Coordinates | Start and end position of this feature, relative to the parent sequence. |
| ORFID | A unique ID given to this feature, primarily for internal use. Only applies to features that are genes. |
| Strand | Specifies if the feature was found in the forward (1) or backward (-1) direction. Only applied to features that are genes. |
| Accession | ID/accession for the reference sequence that had the best alignment (by e-value) with this feature's translated sequence. |
| E_val | The e-value score for the best alignment for this feature. |
| Description | A description of this putative feature, parsed from the defline of best aligned reference sequence. |
| Sequence | The (translated) amino acid sequence for this feature. |
| Bitscore | The bitscore for the best alignment for this feature. |
| Rawscore | The raw score for the best alignment for this feature. |
| Aln_len | The length of the best scoring alignment, in base pairs. |
| Pident | The fraction of identical positions in the best alignment. |
| Nident | The number of identical positions in the best alignment. |
| Mismatch | The number of mismatched positions in the best alignment. |
| Positive | The number of positive-scoring matches in the best alignment. |
| Gapopen | The number of gap openings. |
| Gaps | Total number of gaps in the alignment. |
| Ppos | Percentage of positive scoring matches. |
| Qcovhsp | Query coverage per HSP. That is, the fraction of the query (this feature's translated amino acid sequence) that was covered in the best alignment. |
| Contig_filename | The input data (genomic sequence(s)) file path. |
-
Results/CRISPR/*_filtered/*.png: The visualizations of all predicted
CRISPR array, as shown below: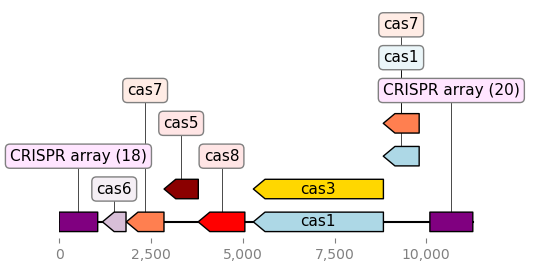
License
MGCA is free software, licensed under GPLv3.
Feedback and Issues
Please report any issues to the issues page or email us at liaochenlanruo@webmail.hzau.edu.cn.
Citation
If you use this software please cite: Hualin Liu. MGCA: microbial genome component and annotation pipeline. Avaliable at GitHub https://github.com/liaochenlanruo/mgca
Updates
V0.0.0
- The MGCA was born.




